Abstract
The prognosis for patients with multiple myeloma (MM) is dependent on a variety of measures including treatment received, clinical factors, and genomic characteristics, with several specific genomic features predicting for shorter progression free survival (PFS). Studies have been conducted to identify biomarkers using gene expression profiles and copy number variation (CNV) data. However, no study to date has integrated genomic data from a systems view, incorporating interactions between biomarkers in a network. We apply an innovative geometric network analysis that integrates complex interactions to characterize patterns of biological behavior not captured by individual genomic events. The methodology is mathematically well-defined and has no fitting parameters. We hypothesized that such a systems mathematical approach applied to gene interaction networks may delineate biologically relevant MM subtypes and potential new therapeutic targets.
We overlaid RNA-Seq and CNV data from the MMRF CoMMpass dataset (IA19) on a gene interactome derived from the Human Protein Reference Database (HPRD) using a novel graph metric of network robustness - Ollivier-Ricci curvature (ORC). ORC considers both local and global connectivity in assessing the robustness of each pathway characterized by feedback loops in a network. Hierarchical agglomerative clustering of the resulting ORC matrix computed for each omics-dataset was performed to identify subtypes based on the patterns of curvature, with the optimal number of clusters determined by silhouette score. Survival analysis for progression free survival (PFS) was performed employing Kaplan-Meier and log-rank tests. A differential gene expression analysis between high and low risk groups, as identified by the gene expression data, was then carried out using raw read counts with DESeq2. Further downstream bioinformatic analysis was conducted using MetaCore software.
Both RNA-Seq and CNV data were available in 659 patients. Gene product interactions were determined by protein-protein interactions derived from HPRD, resulting in a network with 8,468 nodes and 33,695 edges. The ORC analysis using RNA-Seq data resulted in 6 clusters, and Kaplan-Meier analysis showed statistical significance in PFS with log-rank p=0.0016. Clusters predicting the best and worst PFS were associated with specific genomic features including CCND1 and MAF/MAFB translocations, respectively (each p<0.05, Fisher's exact test). Considering the central role of CCND1 in MM biology, we repeated our analysis with t(11;14) patients excluded, with the resulting clustering remaining significant for predicting PFS.
The ORC analysis of CNV data on HPRD resulted in 8 clusters, and Kaplan-Meier analysis showed statistical significance in PFS with log-rank p=0.0082. Genomic features associated with the clusters included hyperdiploidy and CCND1 (longest PFS), and MAF-translocations and APOBEC-activity (shortest PFS) (each p<0.05, Fisher's). The worst PFS cluster had a higher prevalence of chromothripsis (p<0.0001, Fisher's) than other clusters, known to be an independent predictor for MM survival. Considering the impact of hyperdiploidy on CNV in MM, analyzing hyperploid and non-hyperdiploid patients separately was still significant for PFS prediction.
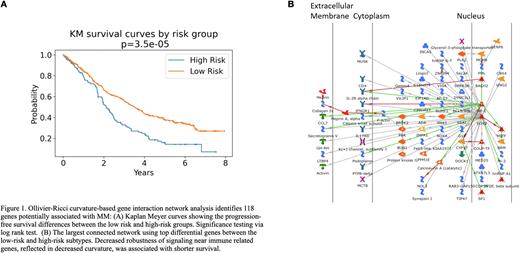
After DESeq2 analysis to perform differential gene expression analysis between the high and low risk groups, the Benjamini-Hochberg method was applied to adjust the resulting p-values, producing false discovery rate (FDR). A list of 118 genes derived with FDR < 0.05 and |fold change| > 3.5 was input into MetaCore to identify biological correlates associated with the risk of PFS. Key implicated biological processes included cell cycle progression, inflammation and immune response.
In this study, we applied the geometric network analysis tool ORC to multi-omics data in MM represented as biological networks to identify individuals at high risk of short PFS and relevant biological correlates. One hundred and eighteen key genes associated with MM prognosis were identified. Decreased robustness of signaling near immune-related genes was associated with shorter survival. For both RNAseq and CNV-based data, patient clusters were associated with known predictors of PFS, and remained predictive when removing specific genomic features, indicating that our methods are providing new biological insights.
Disclosures
Usmani:Abbvie, Amgen, BMS, Celgene, EdoPharma, Genentech, Gilead, GSK, Janssen,Oncopeptides, Sanofi, Seattle Genetics, SecuraBio, SkylineDX, Takeda, TeneoBio: Consultancy; Amgen, BMS, Janssen, Sanofi: Speakers Bureau; Amgen, Array Biopharma, BMS, Celgene, GSK, Janssen, Merck, Pharmacyclics, Sanofi, Seattle Genetics, SkylineDX, Takeda: Research Funding. Norton:Martell Diagnostic Laboratories, Inc: Consultancy, Current equity holder in publicly-traded company; Celgene: Consultancy; Cold Spring Harbor Laboratory: Consultancy; Codagenix, Inc: Consultancy, Current equity holder in publicly-traded company; Agenus Inc.: Consultancy; Immix Biopharma, Inc.: Consultancy, Current equity holder in publicly-traded company; QLS Advisors, LLC: Consultancy; Samus Therapeutics LLC: Consultancy, Current equity holder in publicly-traded company.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal